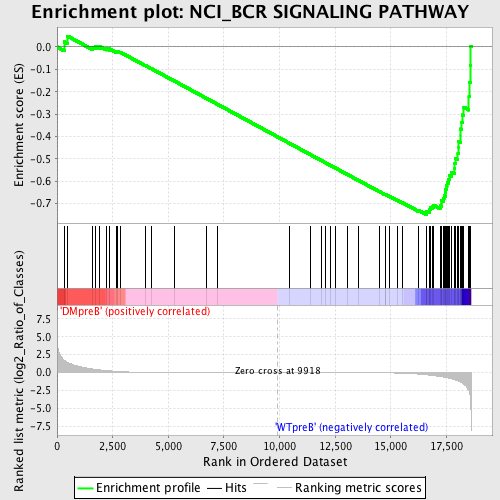
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | NCI_BCR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.7480696 |
| Normalized Enrichment Score (NES) | -1.6055307 |
| Nominal p-value | 0.006437768 |
| FDR q-value | 0.04368458 |
| FWER p-Value | 0.393 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BLNK | 23681 3691 | 325 | 1.702 | 0.0227 | No | ||
| 2 | PPP3CA | 1863 5284 | 472 | 1.412 | 0.0483 | No | ||
| 3 | CAMK2G | 21905 | 1605 | 0.498 | -0.0010 | No | ||
| 4 | MAP3K7 | 16255 | 1728 | 0.446 | 0.0030 | No | ||
| 5 | MAPK8 | 6459 | 1918 | 0.379 | 0.0018 | No | ||
| 6 | MALT1 | 6274 | 2207 | 0.289 | -0.0069 | No | ||
| 7 | CHUK | 23665 | 2349 | 0.253 | -0.0085 | No | ||
| 8 | CSNK2A1 | 14797 | 2669 | 0.175 | -0.0216 | No | ||
| 9 | HRAS | 4868 | 2695 | 0.167 | -0.0190 | No | ||
| 10 | IKBKB | 4907 | 2832 | 0.138 | -0.0231 | No | ||
| 11 | SHC1 | 9813 9812 5430 | 3966 | 0.042 | -0.0831 | No | ||
| 12 | AKT1 | 8568 | 4238 | 0.034 | -0.0969 | No | ||
| 13 | ETS1 | 10715 6230 3135 | 5256 | 0.018 | -0.1513 | No | ||
| 14 | CALM1 | 21184 | 6700 | 0.009 | -0.2289 | No | ||
| 15 | IBTK | 19047 | 7213 | 0.007 | -0.2563 | No | ||
| 16 | LYN | 16281 | 10463 | -0.001 | -0.4314 | No | ||
| 17 | PAG1 | 15376 | 11376 | -0.004 | -0.4805 | No | ||
| 18 | IKBKG | 2570 2562 4908 | 11885 | -0.005 | -0.5077 | No | ||
| 19 | DOK1 | 17104 1018 1177 | 12045 | -0.006 | -0.5162 | No | ||
| 20 | DAPP1 | 11159 6445 6446 | 12268 | -0.007 | -0.5280 | No | ||
| 21 | PIK3R1 | 3170 | 12501 | -0.008 | -0.5403 | No | ||
| 22 | SOS1 | 5476 | 13043 | -0.010 | -0.5692 | No | ||
| 23 | PIK3CA | 9562 | 13536 | -0.014 | -0.5954 | No | ||
| 24 | SH3BP5 | 6278 | 14468 | -0.028 | -0.6449 | No | ||
| 25 | NFKBIB | 17906 | 14759 | -0.037 | -0.6597 | No | ||
| 26 | FOS | 21202 | 14780 | -0.038 | -0.6599 | No | ||
| 27 | VAV2 | 5848 2670 | 14953 | -0.044 | -0.6681 | No | ||
| 28 | MAP2K1 | 19082 | 15278 | -0.061 | -0.6841 | No | ||
| 29 | PTPRC | 5327 9662 | 15526 | -0.080 | -0.6955 | No | ||
| 30 | CALM2 | 8681 | 16243 | -0.203 | -0.7293 | No | ||
| 31 | RAC1 | 16302 | 16592 | -0.304 | -0.7409 | Yes | ||
| 32 | MAP4K1 | 18313 | 16615 | -0.312 | -0.7347 | Yes | ||
| 33 | PPP3CB | 5285 | 16735 | -0.352 | -0.7328 | Yes | ||
| 34 | PPP3CC | 21763 | 16753 | -0.357 | -0.7252 | Yes | ||
| 35 | RASA1 | 10174 | 16776 | -0.365 | -0.7178 | Yes | ||
| 36 | CALM3 | 8682 | 16860 | -0.390 | -0.7130 | Yes | ||
| 37 | RAF1 | 17035 | 16923 | -0.414 | -0.7066 | Yes | ||
| 38 | MAPK14 | 23313 | 17215 | -0.531 | -0.7097 | Yes | ||
| 39 | POU2F2 | 17929 | 17268 | -0.557 | -0.6993 | Yes | ||
| 40 | CARD11 | 8436 | 17271 | -0.561 | -0.6862 | Yes | ||
| 41 | GRB2 | 20149 | 17370 | -0.613 | -0.6770 | Yes | ||
| 42 | MAPK3 | 6458 11170 | 17396 | -0.634 | -0.6633 | Yes | ||
| 43 | JUN | 15832 | 17458 | -0.677 | -0.6506 | Yes | ||
| 44 | CD19 | 17640 | 17470 | -0.683 | -0.6350 | Yes | ||
| 45 | PLCG2 | 18453 | 17511 | -0.711 | -0.6204 | Yes | ||
| 46 | CD22 | 17881 | 17526 | -0.717 | -0.6042 | Yes | ||
| 47 | NFKB1 | 15160 | 17596 | -0.757 | -0.5900 | Yes | ||
| 48 | BTK | 24061 | 17622 | -0.771 | -0.5731 | Yes | ||
| 49 | RELA | 23783 | 17734 | -0.859 | -0.5587 | Yes | ||
| 50 | SYK | 21636 | 17870 | -0.978 | -0.5429 | Yes | ||
| 51 | BCL10 | 15397 | 17878 | -0.984 | -0.5200 | Yes | ||
| 52 | TRAF6 | 5797 14940 | 17908 | -1.020 | -0.4974 | Yes | ||
| 53 | PTEN | 5305 | 18017 | -1.128 | -0.4765 | Yes | ||
| 54 | FCGR2B | 8959 | 18041 | -1.163 | -0.4503 | Yes | ||
| 55 | CD79B | 20185 1309 | 18046 | -1.168 | -0.4228 | Yes | ||
| 56 | PDPK1 | 23097 | 18147 | -1.308 | -0.3973 | Yes | ||
| 57 | NFKBIA | 21065 | 18148 | -1.309 | -0.3663 | Yes | ||
| 58 | CSK | 8805 | 18193 | -1.378 | -0.3361 | Yes | ||
| 59 | CD79A | 18342 | 18218 | -1.423 | -0.3037 | Yes | ||
| 60 | MAP3K1 | 21348 | 18282 | -1.594 | -0.2694 | Yes | ||
| 61 | PTPN6 | 17002 | 18512 | -2.552 | -0.2214 | Yes | ||
| 62 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 18532 | -2.758 | -0.1571 | Yes | ||
| 63 | CD72 | 8718 | 18566 | -3.281 | -0.0813 | Yes | ||
| 64 | MAPK1 | 1642 11167 | 18573 | -3.548 | 0.0023 | Yes |

